Development of regenerative therapeutics involves understanding and application of molecular, cellular and tissue engineering principles. Integrated strategies include biomaterials, therapeutic molecules and stem cells to create bioengineered systems for regenerative medicine. Therefore, understanding fundamental interactions between different components and utilizing these concepts will provide tools to engineer tissue regeneration and develop treatment options for diseases. Major thrust areas are:
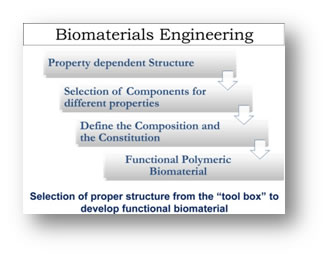
Biomaterials development by combinatorial approach
Therapeutic biomaterials should provide signals to control cell fate in regenerative medicine. Thus, biomaterials with controlled structure and function are essential to induce and maintain cellular functions. We are interested in developing a library of novel polymeric biomaterials by altering the structure and composition. By using a combinatorial approach we want to define structure-property relationship of polymers as therapeutic biomaterial and develop this approach as molecular engineering tool for regenerative applications. Current projects involve developing materials for engineering scaffolds, cell modification and drug delivery.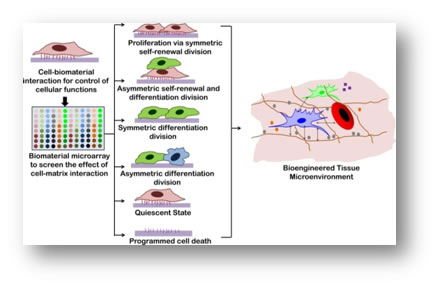
Cell-biomaterial interaction for construction of synthetic tissue microenvironment
Specific interactions between cell and biomaterials are required to control the cellular functions and for development of a cell (or stem cell) niche. These interactions provide rational designs for construction of specific tissue microenvironment for physiological and pathological conditions. We are interested in elucidating the fundamental cross-talk between a cell and material to provide microenvironmental cues and in understanding the role (and interplay) of the cues in controlling cellular functions including stem cell for cell adhesion, proliferation, migration and differentiation. Our goal is to mimic these interactive signaling events between cell and biomaterial for developing a bioengineered niche which can provide signals in with a spatial and temporal control.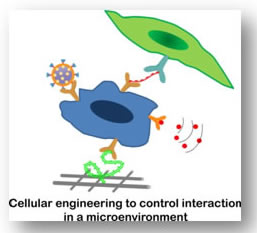
Engineering cell and cellular functions
Programming of cells provides a viable and alternate route to control cell response in a physiological or pathological condition without eliciting an adverse response. Cell-cell and cell-matrix interactions can be controlled by modifying the cells and stem cells to induce or inhibit specific responses. Modification of cells with engineering tools is important aspect to develop regenerative therapeutics. We are interested in engineering cell membranes with biomolecules and polymers to modulate cell targeting and subsequent signaling events in a tissue microenvironment. Additionally, we are aiming to modify cells as a delivery vehicle for transporting therapeutics.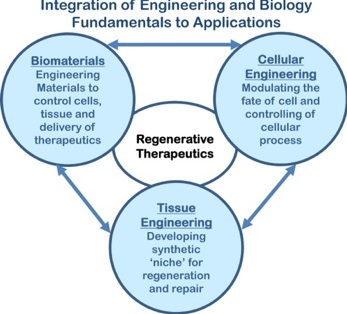
Regenerative therapeutics
The translational aspect of regenerative medicine depends on proper integration of engineering and medicine. Concepts from fundamental science and engineering are converted into reality for specific applications. This hierarchical roadmap is tissue or disease specific and thus requires step-wise approaches. Our current goals are to develop strategies for therapeutic angiogenesis, soft and elastic tissue regeneration and delivery of drugs. We are interested to integrate the different components for effective therapeutic strategies.
Examples of Research Projects
-Studying cell-matrix interactions with in-vitro angiogenesis models, the relationship between synthetic versus natural interfacial properties and mechanical/chemical properties of cells.
-Working with Polyurethanes and functionalize different type of polyurethanes. Studying their rheology and cellular interaction in both 2D and 3D surfaces.
-Researching various polyurethanes affect on the osteogenic differentiation of mesenchymal stem cells. This includes work with in both 2 and 3 dimensions.
-Studying the morphology and behavior of endothelial cells on polyurethanes and how the biphasic organization of the polyurethanes influences the formation of these tube networks.
-Research involving endothelial cells interactions with 3 dimensional networks.
 Biomaterials and Regenerative Therapeutics Laboratory
Biomaterials and Regenerative Therapeutics Laboratory