INTRODUCTION
Novel high-throughput genomic and proteomic tools are allowing the integration of information from a range of biological assays into a single conceptual framework. This framework is often described as a network of biochemical reactions. In such an approach, independent rate constants for a given reaction network are determined using a range of biochemical assays. Algorithms are then designed and implemented in computer programs to integrate and analyze these data. Such an approach has been applied in diverse studies in the fields of signal transduction, protein glycosylation, blood coagulation etc.
To aid the analysis of complex biological reaction networks, we have developed a user-friendly program called “Insilico Biochemical REaction Network Analysis (IBRENA)”. This program is written in MATLABTM (The Mathworks, Inc., Natick, MA) with a Graphical User Interface (GUI). In order to reduce computation time, in some cases, ordinary differential equations (ODEs) are solved using Visual FORTRAN (HP-Compaq V6.6, Palo Alto, CA) with integrated IMSL library (Visual Numerics Inc., Houston, TX). This reduces the computational time required for time-consuming sensitivity analysis (described in Chapter 4 of the manual). This application can be deployed on a PC using Windows.
IBRENA can be installed either as a stand-alone application or it can be used in the MATLAB environment. Simulation results in both cases can be graphed using IBRENA’s built-in plot functions, or these can be exported in comma delimited files for analysis in Matlab or elsewhere.
- STAND ALONE APPLICATION: The stand alone application is easy to install and use. This does not require installation of either the SBML or SUNDIALS toolbox. All inputs for the stand-alone application are provided using .csv (comma separated value) input files. This partially installed version of the software can handle elemental reactions but not more complex reactions.
- FULL VERSION OF THE APPLICATION: The complete version of IBRENA that is run in the MATLAB environment can handle any general reaction network that is described by SBML (Systems Biology Markup Language) type input file and that can be processed using the SUNDIALS package. This then requires installation of the SBML and SUNDIALS toolboxes to solve arbitrary reactions.
Detailed differences between the stand-alone and full-versions are listed in the manual (see chapter 1)
Overall, IBRENA enables:
* Conversion of .csv format input files to SBML format output.
* Insilico dynamic simulation of reaction networks.
* Identification of critical rate-limiting or regulatory reactions (or species) using sensitivity analysis. Both forward and backword (adjoint) sensitivity analysis can be performed using this software. Output M-files are also generated for both types of sensitivity analysis. These M-files may be used by the user for additional user-specific analysis, if necessary, using the array of functions available in MATLAB.
* Analysis of dynamic simulation or sensitivity coefficient data to decipher the connectivity between the reactions or species. Such analysis is performed using Principal Component Analysis (PCA) and Singular Value Decomposition (SVD).
* Model reduction by combining sensitivity analysis with flux analysis.
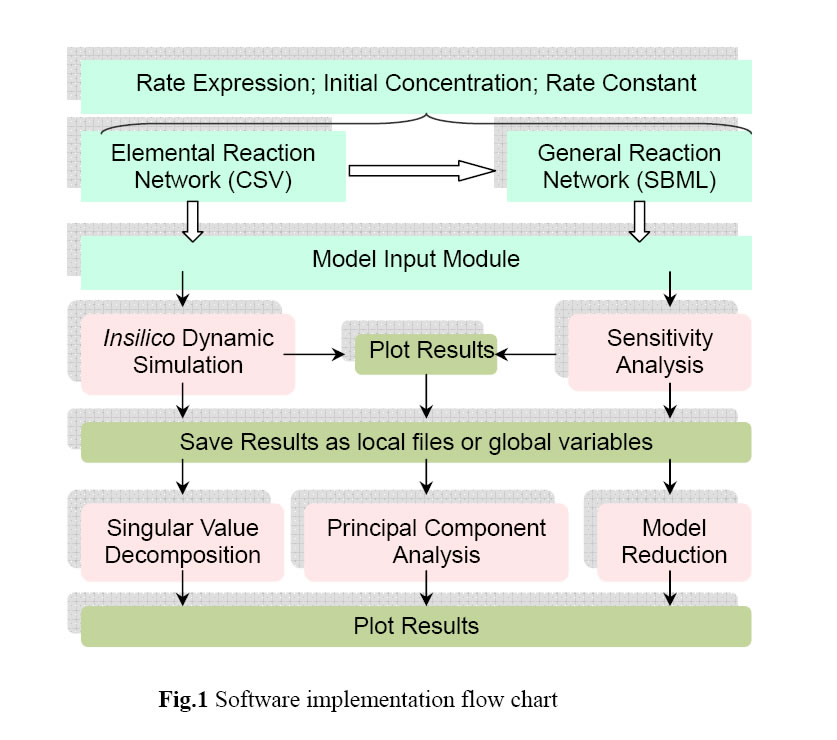
The schematic in Fig.1 illustrates the software implementation scheme. Input for the program can be provided using files in either SBML 2.0/1.0 (Systems Biology Markup Language) or Excel Comma Separated Value (.csv) format. Details on the underlying theory, implementation scheme, and examples are provided in the software manual.
PUBLICATIONS
1.Liu, G. and S. Neelamegham, In silico Biochemical Reaction Network Analysis (IBRENA): a package for simulation and analysis of reaction networks. Bioinformatics, 2008 [PUBMED LINK]
* This paper is the application note that describes the software.
2.Liu, G., M.T. Swihart, and S. Neelamegham, Sensitivity, principal component and flux analysis applied to signal transduction: the case of epidermal growth factor mediated signaling. Bioinformatics, 2005. 21(7): p. 1194-202.[PUBMED LINK]
* This paper provides detailed analysis of EGF-induced signaling network. The SBML and CSV files for this reaction network are provided in IBRENA package as test files.
INSTALLATION
Please see installation instruction in the manual (Chapter 1). The manual can be downloaded below.
DOWNLOAD
Manual and application package can be downloaded from
the links below
Manual.pdf
IBRENAStandalone.exe (120MB
Self-extracting Application Package)
IBRENAFullVersion.exe (14MB Self-extracting Application Package)
System Requirement: Windows XP or 2000 based PC,
Pentium 4 or later versions
COMMENTS
Effort has been taken to ensure complete documentation of the program and tests have been performed to test the robustness and accuracy of the code. If you have any questions or comments (including bug reports), please contact us at gangliu@eng.buffalo.edu (Gang Liu) or neel@eng.buffalo.edu (Sriram Neelamegham).

